MMS Verify and Save
Verify
There are 3 verification routines built into MMS: Alignment Target verification, Guide Star verification, and the Slit verification. These are each run when saving or can be run independently.
Verify Alignment Boxes is a routine that can be run independently or when saving. It performs the following checks:
- Failure message if less than three 4×4 arcsec boxes or circles in the 5.6 arcmin diameter circle
- Warning if there are three boxes in 5.6 arcmin diameter but less than 3 in the 5 arcmin diameter circle
Guide Star Verify is a routine that can be accessed through the Guide Star submenu. It confirms that at least 1 object has been selected. It does not check magnitude or whether the object is extended, so the user will need to independently confirm that there is a suitable candidate that meets the guider criteria when selecting. The magnitude information is not passed on in any way to lms, so when the user makes their scripts in the OT it is recommended that they reselect their guide star.
Verify Slits is a recent addition that allows the user to do a quick assessment of the mask slit placement before sending it in for a final verification/check with our mask scientist. This feature is run each time you save your mask, or can be run independently. The routine checks that science slits are properly placed within the FoV, it performs a first order vertical overlap check, and checks if slits may be too close to one another.
When assessing vertical overlap, the routine does not take into account vertically overlapping reference boxes which are a potential exception to this rule. But does try to flag to the user if they may see vertical overlap from multiple slits. If your science will not be directly impacted by any vertical overlap this can be dismissed.
Protip: The terminal will provide feed back any failures or warnings!
Save / Update Revision
For mask submission you mask must have:
Project Name
A minimum of 3 Alignment Boxes
No slits violated the proximity verification, unless otherwise discussed and verified in advance with the mask scientist (slitmasks@lbto.org)
Highly Recommended
At lease one Guide Star
No slit verification violations
Save and Update Revision will create 3 files, all with the same name for the form:
mods.N..lms, mods.N.gbr, mods.N.epsf
where N is a counter that increments by 1 with each mask generated. The Project Name is a user defined name that can up to 8 character long. If no project name is defined, this part of the file name is omitted and the mask will not load into the OT.
Do not rename the MMS files to something different than what MMS produces.
When saving there is often a popup asking which catalog was used for mask development. This information is used to populate the mms header. Make sure to select a catalog or hit ok to ensure your mask is saved.The mms file contains the information for telescope pointing and instrument rotation, all slit parameters, including slit center coordinates in WCS units (not for the 6 reference apertures) and in mask coordinates (mm x and y) and Alignment boxes, and the WCS coordinates of the Guide Stars. This file can be reloaded to restore the MMS session. It is ingested by the OT for script generation.The gbr file contains the information required for cutting the mask and the slits, including the mask contour, the holes for mounting of the mask, and the mask ID. The epsf file contains several known bugs including the inability to display the file number and curved slits. (see known bugs).When submitting, include the mms and gerber file. If referring to a previously cut mask it is often best to resend the mms files for verification. The MASKID is the number cut at the top of the mask and can be found in the header of your MMS file:
Gerber viewer
We recommend gerbv for viewing your mask gerber files. This will allow you to measure physical sizes and see what will be cut by our laser milling machine. Gerbv is a free open source gerber file viewer.
MAC OS X: Macports should be installed from your installation and configuration of skycat. This can be used to also install your gerbv.
Linux: Install instructions for linux and other OS can be found here: http://gerbv.geda-project.org/
Checking for Slit Overlap
- download slitmap3.py
- edit the first line (starting with #!) to point to your python executable
- chmod +x slitmap.py to insure it is executable
- then run it:
- ./slitmap.py yourmask.mms [MODS1B|MODS1R|MODS2B|MODS2R]
- Note that you must run this for each channel (MODS1B, MODS1R, MODS2B, or MODS2R) since the transform is different for each.
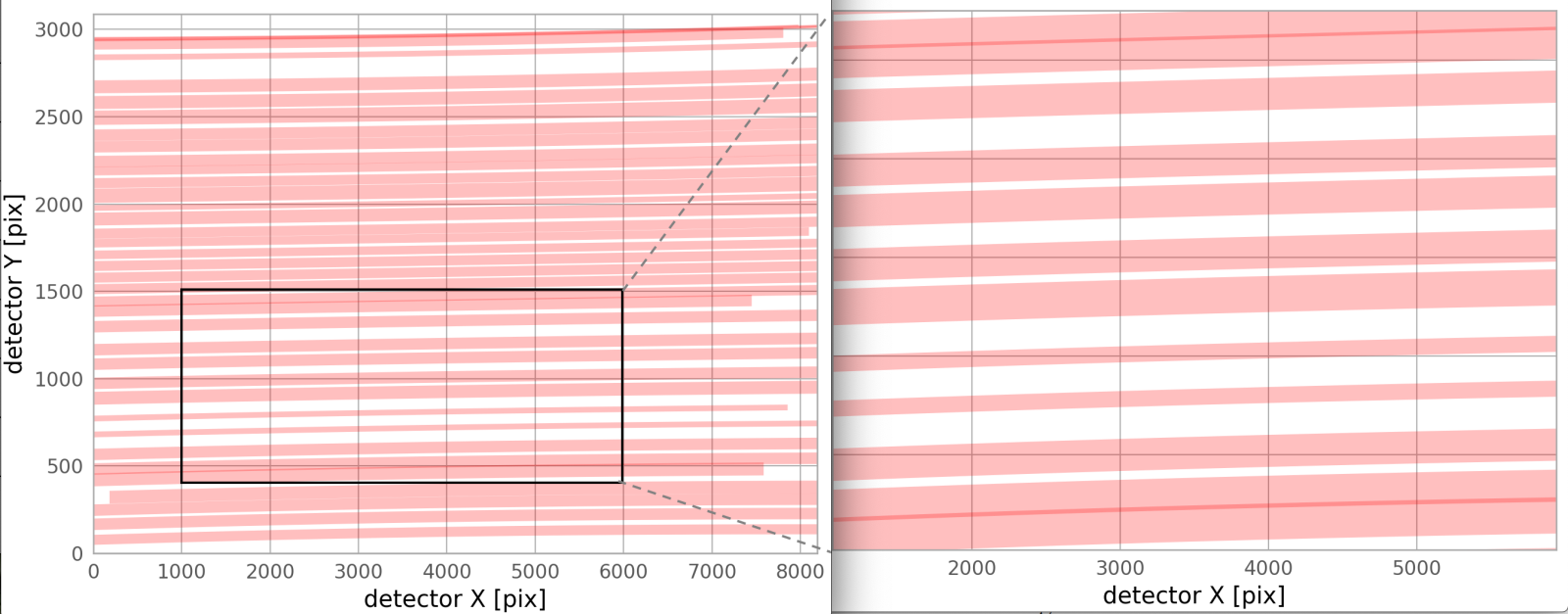
- The output plot will look something like the following, which is a prediction for where the traces for a given mms file will fall on the MODS2R full-frame, 8192 x 3088 pixel detector. Currently, the predicted traces simply cover same the wavelength range, 3200-11500 angstroms, for both channels. There are slight overlaps between a few traces, as the figure shows.